

|
GLOB
Nitrogen gain and loss in changing coastal lagoon ecosystems of the Baltic Sea |
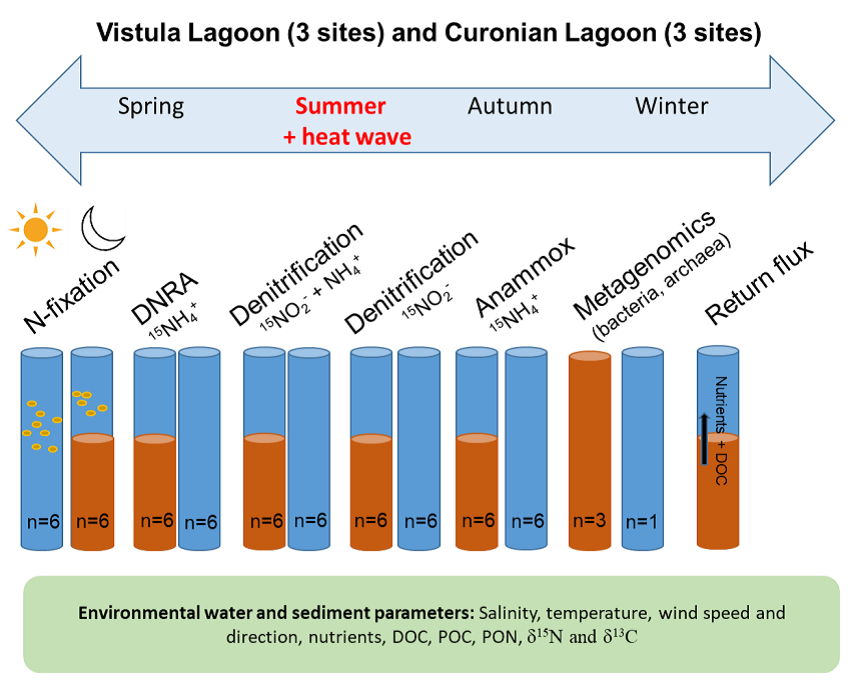 Conceptual overview of the experimental designs assessing N-gain and N-loss processes in the coastal lagoons |
Research methodology
In GLOB, we will use a unique combination of methods including a quantification of nutrient cycling pathways (Biogeochemical), such as fixation and reduction, analysis of microbial taxonomic and functional biodiversity (Functional genomics), as well as assessment of ecosystem functioning (Ecological analysis). All field campaigns will be conducted seasonally (winter, spring, summer and autumn) in two dominant macro-areas of the lagoons. We will also monitor N inputs via main tributaries (Nemunas, Pasleka) to estimate loads. Additionally, a set of experiments will be carried out to estimate marine heat waves' effects on N cycling processes. For this purpose, we will collect additional samples (between June and September) to simulate increased temperature effects (+4, +8 and +12 °C) on selected processes. All samplings will be carried out from boats using a Go Flow sampler for water sampling and a hand-handled sediment corer for intact sediment core collection. The samples for measuring rates will be taken in multiple replicates to elucidate variability in targeted processes. Collected samples will be immediately transported to the laboratory under controlled conditions for further treatments and experimental activities. In addition, state-of-the-art CTD probes will be used for fine-scale water column profiling. Dissolved nutrient analysis will be carried out on a segmented continuous flow analyzer using spectrophotometric methods.Nutrients (POC, PON and their isotopic composition) in particulate matter from in situ sampling and experiments will be measured via an elemental analyzer coupled to an isotope ratio mass spectrometer (IRMS). Samples for pelagic and benthic N₂ fixation rates will be ¹⁵N₂-enriched, and isotope incorporation rates in microorganisms’ biomass will later be measured with an elemental analyzer coupled to an IRMS. The ¹⁵N-atom % in the dissolved N₂ pool from the fixation experiments will be quantified using a high-precision membrane inlet mass spectrometer. Samples for N reduction processes (denitrification, DNRA and anammox) will be incubated in another set of microcosms as for fixation experiments. The production of ²⁹/³⁰N₂ (to calculate denitrification and anammox rates) and ¹⁵NH₄⁺ (to calculate DNRA rates) will be analyzed using gas chromatography coupled to an IRMS. Samples for ¹⁵NH₄⁺ production will be analyzed by the same technique after the conversion of NH₄⁺ to N₂ by the addition of alkaline hypobromite. Taxonomic composition of microorganisms involved in the N-cycle will be defined based on 16S rRNA gene sequences amplified from extracted nucleic acids, targeting the V3 region of the gene sequence. Next-Generation Sequencing (NGS) will be carried out on an Illumina MiSeq according to protocol. Sequence data will be denoised using the DADA2 pipeline and the remaining amplicon sequence variants (ASVs) will be classified against the SILVA 138 database. Microbiome (genes + microbiota) associated with benthic C and N cycling will be identified using shotgun-based metatranscriptomic analysis on Illumina NextSeq. The sequences will be classified against the NCBI NR protein database using DIAMOND software. MEGAN will be used to link NCBI NR accession identifiers to NCBI taxonomy and KEGG functional classifications. Measurements will be conducted according to state-of-the-art procedures. Certified reference materials and standards will be applied to verify quality.